|
|
6 months ago | |
|---|---|---|
| DrugCombPred_DrugCombDB | 6 months ago | |
| DrugCombPred_O'Neil | 6 months ago | |
| Images | 6 months ago | |
| .gitignore | 6 months ago | |
| Readme.md | 6 months ago | |
| requirements.txt | 6 months ago | |
Readme.md
DrugCombPred - Drug Combination Synergy Prediction
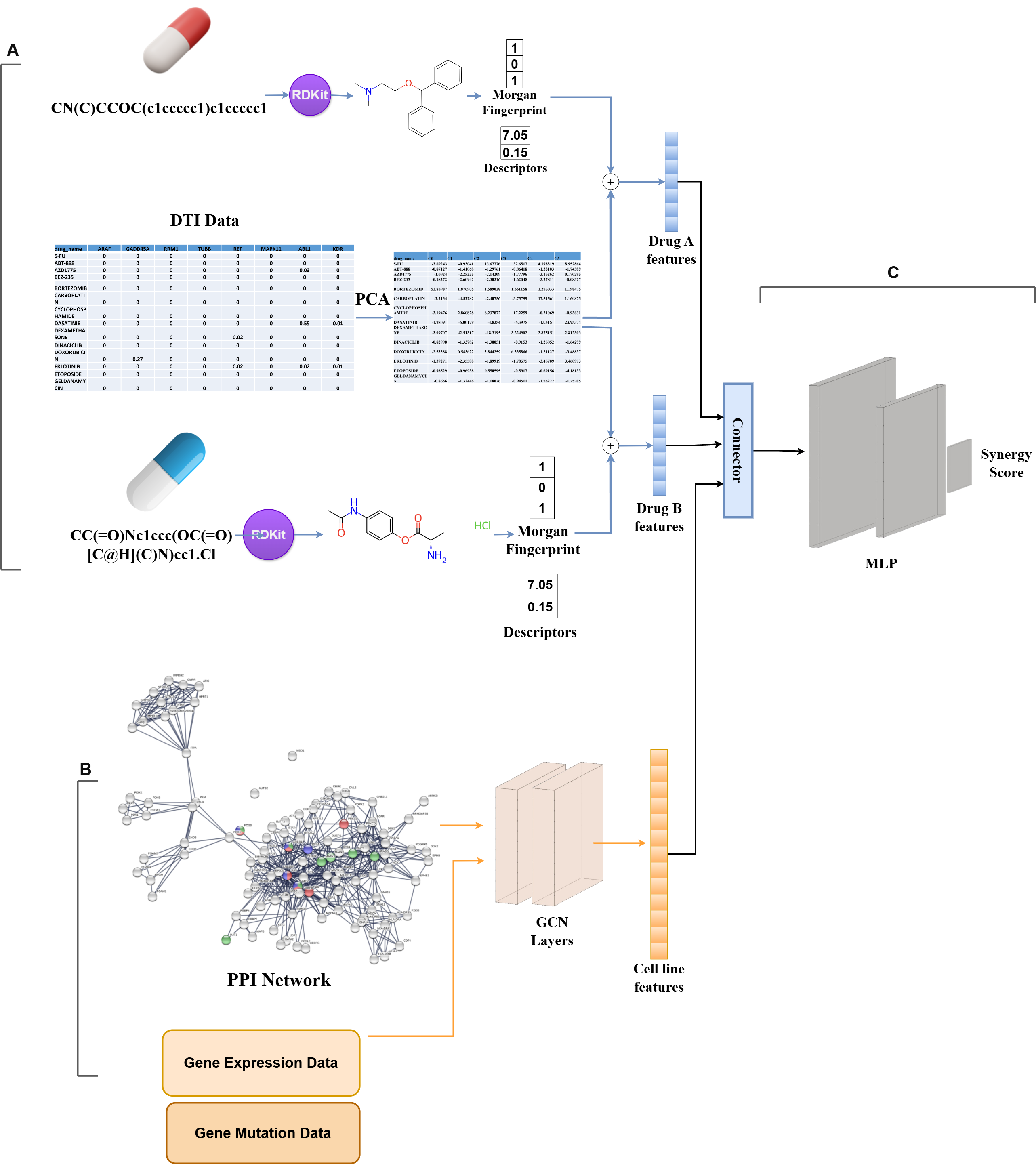
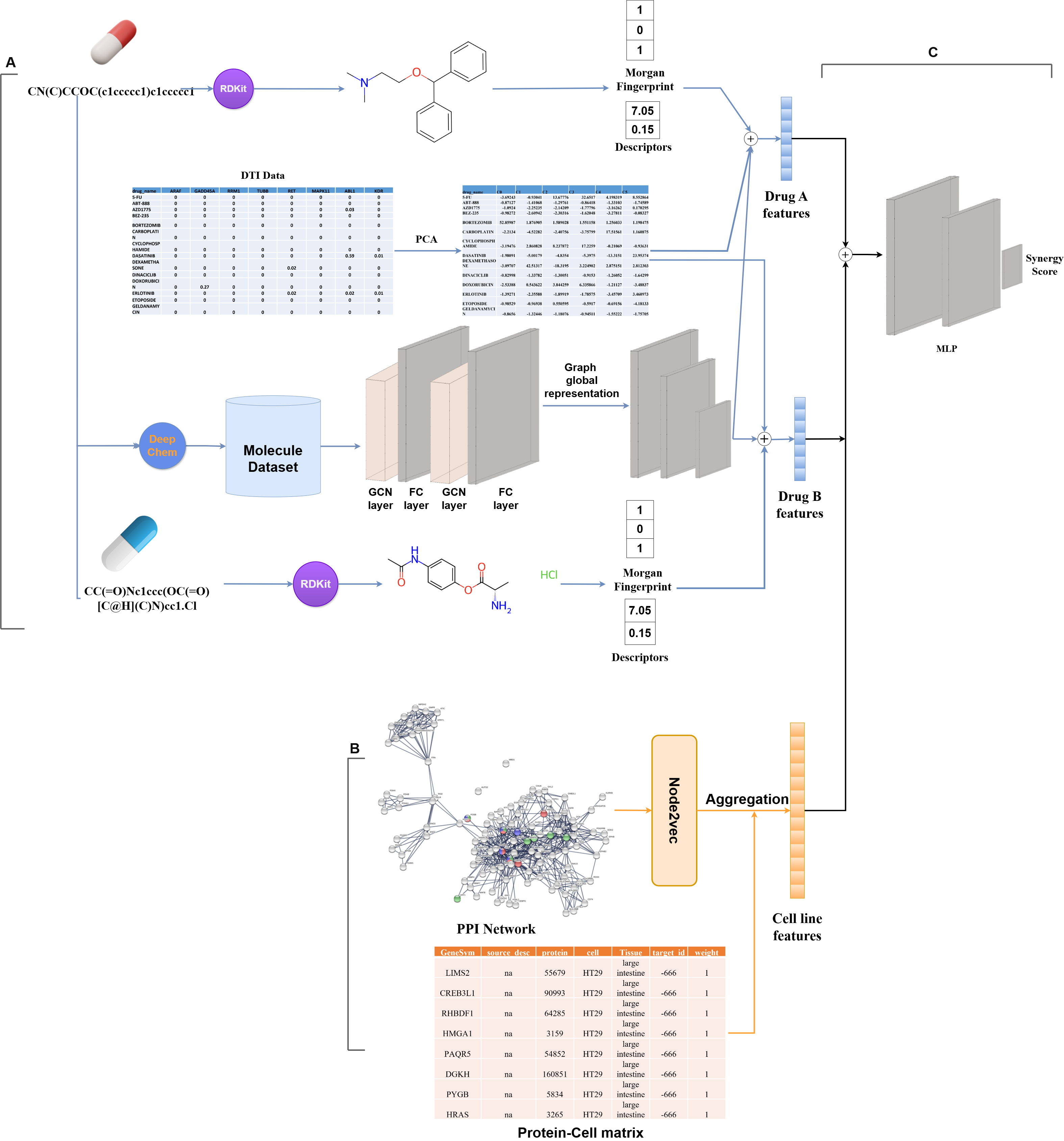
This project focuses on predicting drug combination synergy using deep learning. Combination therapies are a promising strategy in treating complex diseases like cancer, and accurate prediction of drug synergy can help identify effective combinations faster and more cost-effectively. Our models aim to support this effort by learning from large-scale pharmacogenomic datasets.
Two datasets are supported:
- DrugCombDB
- O’Neil et al. (2016)
Each dataset has its own folder and model implementation. The main training script is:
predictor/cross_validation.py
📂 Folder Structure
drugcombpred/
├── DrugCombDB/
│ ├── drug/
│ ├── cell/
│ └── predictor/
│ └── cross_validation.py
├── ONeil2016/
│ ├── drug/
│ ├── cell/
│ └── predictor/
│ └── cross_validation.py
🧠 Project Overview
The models process chemical structures (using RDKit), drug-target interactions, and cell line features (from gene expression and mutation data). Drug features are combined and passed into a neural network to predict synergy scores. Two architectures are used:
⚙️ Requirements
- Python 3.7+
- PyTorch
- CUDA (optional, for GPU support)
Install dependencies:
pip install -r requirements.txt
🚀 Running the Models
Both models use the same script interface: cross_validation.py
Arguments
| Argument | Description | Default |
|---|---|---|
--epoch |
Number of training epochs | 500 |
--batch |
Batch size | 256 |
--gpu |
GPU device ID to use (None for CPU) |
None |
--patience |
Early stopping patience | 100 |
--suffix |
Suffix for output folder | Current timestamp |
--hidden |
Hidden layer sizes (list of ints) | [2048, 4096, 8192] |
--lr |
Learning rate(s) to try (list of floats) | [1e-3, 1e-4, 1e-5] |
Example: Run DrugCombDB Model
cd DrugCombDB/predictor
python cross_validation.py --epoch 300 --batch 128 --gpu 0 --suffix drugcomb_test
Example: Run O’Neil 2016 Model
cd ONeil2016/predictor
python cross_validation.py --epoch 300 --batch 128 --gpu 0 --suffix oneil_test
📁 Output
Results and logs are saved in:
OUTPUT_DIR/cv_<suffix>
Make sure OUTPUT_DIR is defined or configurable in your script.
📚 Citation
If you use this code or data in your research, please cite the original datasets:
DrugCombDB:
Zagidullin et al., Nucleic Acids Research, 2019
https://doi.org/10.1093/nar/gky1144O’Neil et al., 2016:
O’Neil et al., Cell Systems, 2016
https://doi.org/10.1016/j.cels.2016.08.015